Benchmarks¶
Overview¶
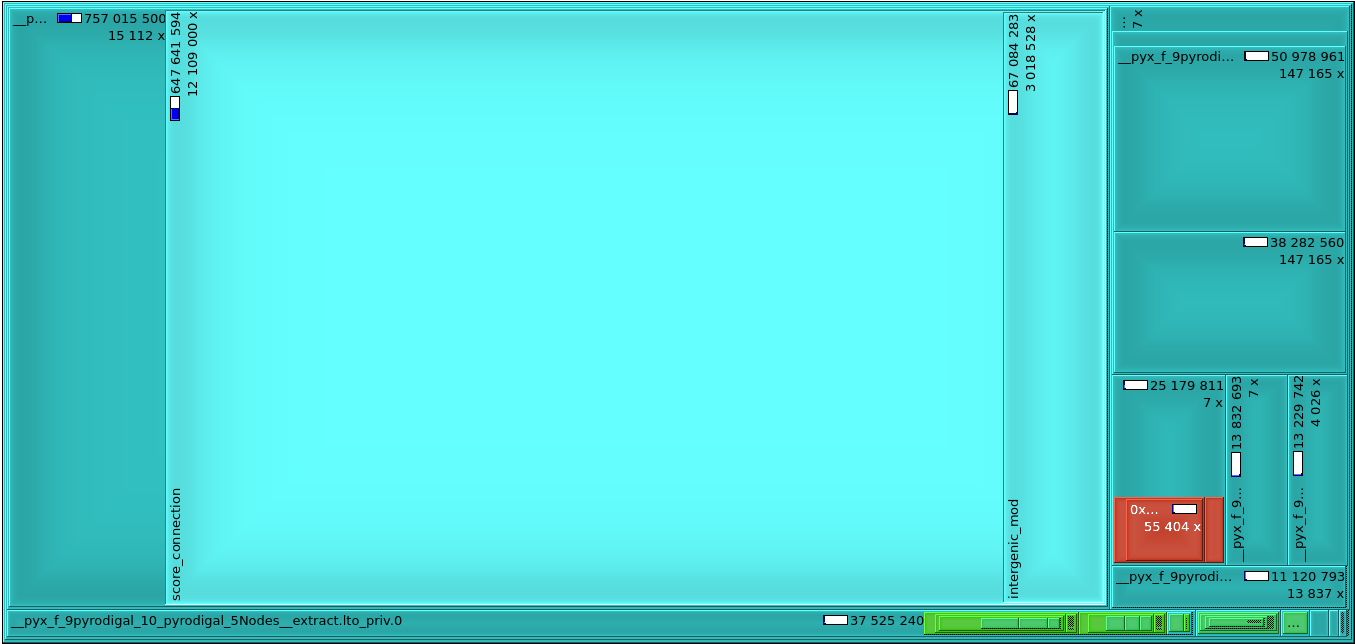
The gene finding in Prodigal is based on dynamic programming, and the performance critical part is actually the node connection scoring step, which attempts to construct the highest-scoring chain of genes inside the input sequence.
Profiling with Valgrind reveals that it represent two-thirds of the total cycle count used for finding genes with the original Prodigal code (647M cycles out of 965M on an example sequence), as seen below:
In Pyrodigal, the connection scoring step is being accelerated with a pre-filtering step so that invalid connections can be skipped. This pre-filtering step can be computed using SIMD instruction, processing several nodes at once. Platform-accelerated code is available for MMX, SSE, AVX, and NEON instruction sets. In addition, a “generic” implementation of the pre-filter is available in plain C for testing purposes. The pre-filtering can be disabled, in which case Pyrodigal will fall back to using the original Prodigal code.
Data & Hardware¶
All benchmarks were done using 50 bacterial genomes of various size and content from the proGenomes database. x86-64 Benchmarks were run on an Intel NUC i7-10710U CPU @ 1.10GHz. Arm benchmarks were run on a Raspberry Pi 4 Model B ARM Cortex-A72 @ 1.5GHz.
Connection Scoring¶
This benchmark evaluates the time needed for scoring all the nodes from a sequence
using either the pre-filter implemented with SIMD (avx, sse, mmx, neon),
the generic pre-filter (Generic) or no pre-filter at all (None).
x86-64¶
Arm¶
Single Run¶
This benchmark evaluates the time needed for an entire genome to be processed
(both training and gene prediction) by Pyrodigal depending on the use of
a SIMD pre-filter (avx, sse, mmx, neon), the generic pre-filter (Generic),
no pre-filter at all (None); or the time taken by Prodigal (Prodigal).